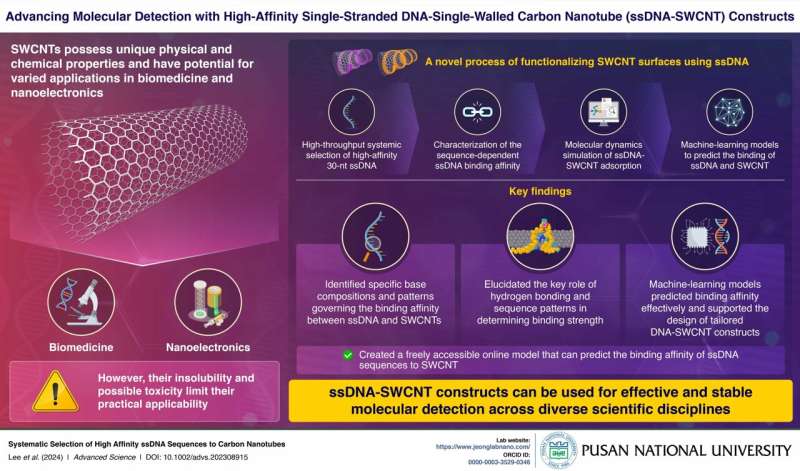
Single-walled carbon nanotubes (SWCNTs) have emerged as promising candidates for purposes in biotechnology and nanoelectronics because of their distinctive bodily and chemical properties. Regardless of their potential, challenges like insolubility and toxicity have hindered their widespread use. Prior research have investigated various methods to functionalize and modify the surfaces of SWCNTs to beat these challenges.
In a brand new research, researchers from the Pusan Nationwide College led by Professor Sanghwa Jeong, Assistant Professor within the Faculty of Biomedical Convergence Engineering, have tried to fill this hole. This research has gone past standard strategies by using high-throughput screening strategies to elucidate the connection between DNA sequences and their binding affinity to carbon nanotubes. It centered on optimizing the binding affinity and stability of those constructs by means of superior sequence design and molecular dynamics simulations.
This research is printed in Superior Science.
Discussing the background of their research, Dr. Jeong explains, “Researchers have been exploring various strategies to engineer SWCNT surfaces to overcome the challenges of limited applications owing to insolubility and potential toxicity. One promising approach is the use of single-stranded DNA (ssDNA) as a wrapping surfactant for SWCNTs.”
The researchers employed a rigorous methodology to make sure exact characterization and optimization of single-stranded DNA (ssDNA)-SWCNT complexes. Initially, a various random 30-nucleotide (nt) ssDNA library underwent iterative rounds of screening to establish high-affinity sequences.
Computational modeling, notably molecular dynamics simulations, supplied insights into the structural dynamics of the SWCNT constructs. Moreover, the researchers used a number of machine-learning fashions to grasp the sample of sequences that have an effect on binding affinity. They’ve efficiently created a freely accessible on-line service that predicts the binding affinity of ssDNA sequences to SWCNTs. These built-in approaches not solely validated the experimental findings but additionally guided the design of high-performance ssDNA-SWCNT constructs.
The findings revealed vital developments within the stability and performance of ssDNA-SWCNT complexes. Excessive-affinity 30-nt ssDNA sequences, wealthy in adenine and cytosine, exhibited superior binding power, validated by means of surfactant displacement experiments.
Molecular dynamics simulations highlighted the formation of secure intramolecular hydrogen bonds close to the SWCNT floor, underscoring their enhanced structural integrity. The machine-learning fashions successfully predicted the binding affinities of ssDNA sequences, additional supporting the design of the tailor-made ssDNA-SWCNT constructs.
Furthermore, the research demonstrated notable enhancements within the resistance of those complexes to enzymatic degradation in comparison with free ssDNA, making them extremely appropriate for long-term organic purposes.
In conclusion, the event of high-affinity ssDNA-SWCNT constructs marks a big development in nanobiotechnology. The distinctive traits of ssDNA-SWCNTs make them splendid candidates for cell or tissue-specific drug supply programs in addition to the event of high-performance nano-electronic gadgets.
Dr. Jeong concludes, “Our study not only makes a substantial contribution to our understanding of the interplay between ssDNA and SWCNTs but also offers practical avenues for harnessing these interactions in a wide range of advanced technologies. In the future, developing nanomaterials and devices with enhanced stability will show promise in driving innovation in nanoelectronics and biotechnology.”
Extra info:
Dakyeon Lee et al, Systematic Collection of Excessive‐Affinity ssDNA Sequences to Carbon Nanotubes, Superior Science (2024). DOI: 10.1002/advs.202308915
Supplied by
Pusan Nationwide College
Quotation:
Researchers discover interaction between high-affinity DNA and carbon nanotubes (2024, July 25)
retrieved 25 July 2024
from https://phys.org/information/2024-07-explore-interplay-high-affinity-dna.html
This doc is topic to copyright. Other than any truthful dealing for the aim of personal research or analysis, no
half could also be reproduced with out the written permission. The content material is supplied for info functions solely.

